The chemical degradation method
Maxam–Gilbert sequencing is a method of DNA sequencing developed by Allan Maxam and Walter Gilbert in 1976–1977. It’s based on nucleobase-specific partial chemical modification of DNA and subsequent cleavage of the DNA backbone at sites adjacent to the modified nucleotides. The chemical degradation method (Maxam–Gilbert sequencing) along with the enzymatic chain termination method (Sanger et al., 1977) were the first widely adopted methods for DNA sequencing and represented the first generation of DNA sequencing methods. As compared to Maxam–Gilbert sequencing, the Sanger chain termination method generates more easily interpreted raw data and has become the most widely used. On the other hand, Maxam–Gilbert method is no longer in common use for routine sequencing.
- Maxam–Gilbert sequencing, developed by Allan Maxam and Walter Gilbert, was published in 1977.
- Despite its initial success, it fell out of favour due to technical complexity, use of hazardous chemicals, and difficulties in scale-up.
- Maxam and Gilbert’s 1977 paper received the Citation for Chemical Breakthrough Award from the Division of History of Chemistry of the American Chemical Society in 2017.
- The award recognized their significant contribution to DNA sequencing as it paved the way for automated high-throughput DNA sequencing possibilities.
Overview of Maxam–Gilbert sequencing
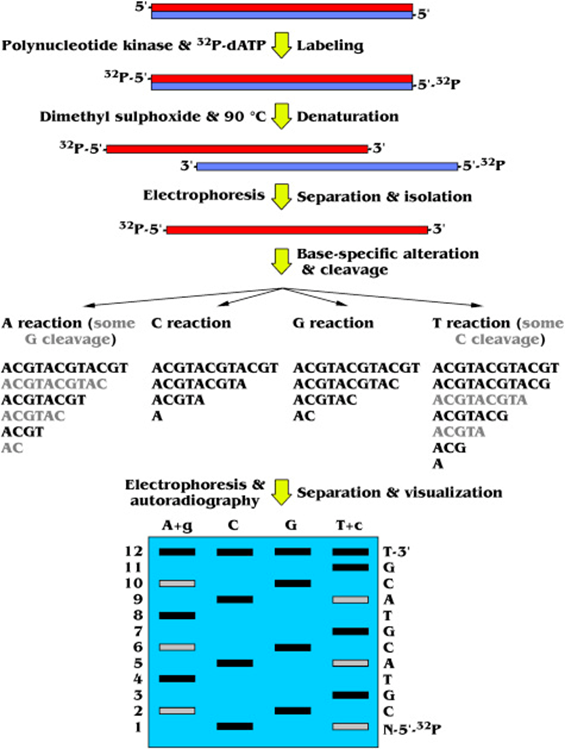
- The procedure involves the radioactive labeling of the 5′-P ends of double-stranded DNA (dsDNA) with 32P-dATP using polynucleotide kinase.
- Then, the DNA is denatured with DiMethyl SulfOxide (DMSO) at 90°C and the resulting ssDNA molecules are segregated via electrophoresis.
- Nitrogenous base-specific reactions are carried out to modify the Adenosine (A), Cytidine (C), Guanosine (G) and Thymidine (T) residues, allowing the chemical cleavage of the ssDNA at the 5′-P side of such positions.
- The A and T reactions also generate some minor G and C cleavage, respectively, which should be taken into account (showing as a weaker signal later on).
- The subsequent polyacrylamide gel electrophoresis and autoradiography allows the ssDNA fragment separation by size and the detection of the radiolabeled DNA band pattern on an x-ray film that encodes the DNA sequence.
An example Maxam–Gilbert sequencing reaction. Cleaving the same tagged segment of DNA at different points yields tagged fragments of different sizes. The fragments may then be separated by gel electrophoresis

Maxam–Gilbert sequencing Procedure
- Radioactive Labelling: One 5′ end of the DNA fragment is labelled with radioactivity (typically gamma-32P ATP).
- Purification: The labelled DNA is purified for subsequent chemical treatments.
- Chemical Treatment: Four reactions (G, A+G, C, C+T) are performed to generate breaks at specific nucleotide bases.
- Purines (A+G) are depurinated using formic acid.
- Guanines (and partially adenines) are methylated by dimethyl sulfate.
- Pyrimidines (C+T) are hydrolyzed using hydrazine.
- Thymine reaction is inhibited in the C-only reaction by adding salt (sodium chloride).
- Cleavage: The modified DNAs are cleaved at the position of the modified base using hot piperidine ((CH2)5NH).
- Fragmentation: Fragments are generated from the radiolabeled end to the first “cut” site in each molecule.
- Electrophoresis: Fragments from the four reactions are electrophoresed in denaturing acrylamide gels for size separation.
- Visualization: The gel is exposed to X-ray film for autoradiography, producing dark bands indicating the location of radiolabeled DNA molecules.
- Inference of Sequence: The sequence is inferred based on the presence or absence of certain fragments
Advantages
- Direct sequencing without cloning
- Simple and fast workflow
- Low amounts of DNA needed
- Can sequence short fragments more precisely
Disadvantages
- Technically complex multi-step process
- Uses hazardous chemicals like hydrazine and formic acid
- Low throughput and difficulties scaling up
- Radioactive labeling generates toxic waste
- Less accurate for long sequences
Applications of Maxam–Gilbert sequencing
- Early small-scale research sequencing
- Sequencing short DNA fragments
- DNA footprinting for identifying protein binding sites
- Detecting cleaved bases damaged by mutations or chemicals
- Analyzing chemical modification of bases like methylation
Challenges of Maxam–Gilbert sequencing
- Technical complexity, hazardous chemicals, and challenges in scaling up led to the decline in popularity.
- The Sanger method’s improvements, such as the chain-termination method, contributed to the reduced use of Maxam–Gilbert sequencing in molecular biology kits
References
- Maxam, A. M., & Gilbert, W. (1977). A new method for sequencing DNA. Proceedings of the National Academy of Sciences, 74(2), 560–564.
- Dorado, S. Gálvez, H. Budak, T. Unver, P. Hernández, Nucleic-Acid Sequencing, Encyclopedia of Biomedical Engineering, Elsevier, 2019, Pages 443-460, ISBN 9780128051443,
- https://en.wikipedia.org/wiki/Maxam–Gilbert_sequencing
- David W. Mount (2001). Bioinformatics: Sequence and Genome Analysis. 1st ed, Cold Spring Harbor Laboratory Press. 98-101.
- Jeremy M. Berg et al., (2002) Biochemistry. 5th edition. New York: W H Freeman. Sections 5.5 and 7.6.
- Graziano Pesole; Cecilia Saccone (2003). Handbook of comparative genomics: principles and methodology. New York: Wiley-Liss. p. 133. ISBN 0-471-39128-X.
- American Chemical Society (2017): Chemical Breakthrough Award recognizing 1977 DNA sequencing paper.